Code
update.packages(checkBuilt = TRUE)R
Rtry() StatementsR
//n with a spaceR on a Network Drive“
Rwill always be arcane to those who do not make a serious effort to learn it. It is not meant to be intuitive and easy for casual users to just plunge into. It is far too complex and powerful for that. But the rewards are great for serious data analysts who put in the effort.”
— Berton Gunter R-help August 2007 (archived at https://perma.cc/KY9N-2FTT)
“Evelyn Hall: I would like to know how (if) I can extract some of the information from the summary of my nlme.
Simon Blomberg: This is
R. There is no if. Only how.”
— Evelyn Hall and Simon ‘Yoda’ Blomberg, R-help April 2005 (archived at https://perma.cc/KY9N-2FTT)
R
Here are a slew of resources for learning base R (in addition to the documents in the lab’s Primer Articles folder):
R for Data Science book: https://r4ds.hadley.nz
r4ds.tutorials package: https://ppbds.github.io/r4ds.tutorials/
R: https://www.statmethods.net (archived at https://perma.cc/SPQ2-DJKM)
R
Codeacademy has free online courses:
R: https://www.sharpsightlabs.com/blog/are-you-fluent-r/ (archived at https://perma.cc/RWL5-GTWV)R skills: https://www.pluralsight.com/search?q=R (archived at https://perma.cc/U4ZZ-UE4X)Cookbook for R to find solutions to common tasks and problems: http://www.cookbook-r.com (archived at https://perma.cc/5ERJ-VRFR)R Graph Gallery to find examples of various graphs: https://r-graph-gallery.com
Coursera courses on R: https://pairach.com/2012/12/22/learn-to-use-r-for-free-with-coursera/ (archived at https://perma.cc/XJ5U-5E4W)R: https://www.r-bloggers.com/moocs-and-courses-to-learn-r/ (archived at https://perma.cc/MQ25-E3ME)Coursera: https://blog.revolutionanalytics.com/2012/12/coursera-videos.html (archived at https://perma.cc/6FU4-PAQW)RStudio Webinars: https://www.rstudio.com/resources/webinars/ (archived at https://perma.cc/6RNR-98JB)R course: https://www.datacamp.com/courses/free-introduction-to-r (archived at https://perma.cc/6ZM9-37L9)R in a Kinder, Gentler, More Effective Manner: https://github.com/matloff/TidyverseSkeptic (archived at https://perma.cc/QU9F-FBS8)R interactively with swirl: https://swirlstats.com (archived at https://perma.cc/TT9V-U663)learnr package: https://blog.rstudio.com/2017/07/11/introducing-learnr/ (archived at https://perma.cc/XGJ8-EXSN)R-bloggers: https://www.r-bloggers.com (archived at https://perma.cc/EL3X-ZXBB)R)The following are resources for learning statistics using R.
R for Data Science: https://r4ds.hadley.nz
Codeacademy has free online courses:
R: https://learningstatisticswithr.com (archived at https://perma.cc/SY87-5GCX)R: https://www.amazon.com/Discovering-Statistics-Using-Andy-Field/dp/1446200469 (archived at https://perma.cc/9LGU-7ZME)R/statistics through the lens of fantasy football: https://isaactpetersen.github.io/Fantasy-Football-Analytics-Textbook
tidyverse
The following are resources for learning tidyverse, which is a collection of R packages for data management:
If you have R questions, you can ask them in a number of places:
Posit: https://forum.posit.co
StackOverflow: https://stackoverflow.com/questions/tagged/r
Reddit: https://www.reddit.com/r/rstats/
R mailing list: https://stat.ethz.ch/mailman/listinfo/r-help
The following article provides additional resources and good guidance: https://www.r-bloggers.com/where-to-get-help-with-your-r-question/ (archived at https://perma.cc/4RRE-KL33).
When posting a question on forums or mailing lists, keep a few things in mind:
R is free software. People are offering their free time to help. They are under no obligation to help you. If you are disrespectful or act like they owe you anything, you will rub people the wrong way and will be less likely to get help.R: https://stackoverflow.com/a/5963610 (archived at https://perma.cc/PC9L-DQZG). Provide a reprex whenever possible: https://reprex.tidyverse.org.Note: many of these initial setup steps described below are not necessary for general use; many of these steps are necessary only for using lab-related repositories (e.g., to gain API access to export data from REDCap, to use absolute paths rather than relative paths so repos can communicate with each other, etc.).
R (https://www.r-project.org) into a directory that contains no spaces; On PC, change the location from the default C:/Program Files/R/[R-VERSION] (which contains a space) to C:/R/[R-VERSION] (which does not contain any spaces; archived at https://perma.cc/6VMX-3LYX—this is because some packages that require compilation to install cannot read filepaths with spaces; archived at https://perma.cc/XA3V-JTPY); may have to right click and “Run As Administrator”
R was already installed in a directory that contains spaces (e.g., C:/Program Files/R/[R-VERSION]), uninstall R before installing it in a directory that doesn’t contain spacesRStudio Desktop (https://www.rstudio.com/products/rstudio/download/) in the main program files directory; may have to right click and “Run As Administrator”. RStudio is the best available graphical user interface for R.R and RStudio to always run with administrator permissions.
R (C:/R/[R-VERSION]/bin/R.exe) and RStudio (C:/Program Files/RStudio/bin/RStudio.exe). Right-click it to open the contextual menu. Then, click or tap on “Properties”. In the Properties window, go to the Compatibility tab. At the bottom of the window, check the box next to the “Run this program as an administrator” option, and then click or tap on Apply or OK.R packages so you can install packages from source, if necessary (i.e., if package binaries are not available):
git, GitLab, and the GitHub Desktop App in the main program files directory; may have to right click and “Run As Administrator”; For instructions setting up and using GitLab, see here: https://devpsylab.github.io/DataAnalysis/git.html#toBegin
Rprofile.site file in the etc folder of the R installation directory is the code that is run for every user at the beginning each time you load R. We will update the default Rprofile.site file with the lab’s Rprofile.site file so R installs packages in the correct location, sets the default package repository, updates packages, and gives you a fortune cookie. To do this, perform the following steps:
Rprofile.site file in the R installation directory (C:/R/R-[InsertVersionNumber]/etc/Rprofile.site) to be Rprofile_BACKUP.site
Rprofile.site file located in this repository at the following location (https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/R%20Setup%20Files/Rprofile.site), and paste it into the R installation directory (PC: C:/R/R-[InsertVersionNumber]/etc/Rprofile.site; Mac: /Library/Frameworks/R.framework/Versions/[InsertVersionNumber]/Resources/etc/Rprofile.site).Rprofile file in the user’s Documents folder is the code that is run for the particular user at the beginning each time you load R. We will update the default .Rprofile file (if there is one) with the lab’s .Rprofile file so R knows which computer you are using and which path to use (relative to where your R projects are located). To do this, perform the following steps:
.Rprofile template file in this repository at the following location, and make sure to remove anything besides .Rprofile in the filename: https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/R%20Setup%20Files/.Rprofile
.Rprofile file, and revise it with your HawkID.Rprofile file with the local path to the Documents folder for each of the computers you will use to access R (e.g., home computer, work computer, laptop). Make sure to use forward slashes (/), not back slashes (\) in the path.HOME directory. To find the HOME directory, open R and type the following command: Sys.getenv("HOME")—the output of the command is the location of your HOME directory; If this is a lab computer, it may be located here: //home.iowa.uiowa.edu/[user]/Documents. If this is your personal computer, it may be located here: PC: C:/Users/[user]/Documents; Mac: /Users/[user]. Then close R.HOME directory is in a OneDrive folder (or another cloud-based sync folder), you will want to change the directory of your HOME path so that it is not in a OneDrive folder. To do that, open Environment Variables (archived at https://perma.cc/A2E5-B5VA) in Windows. Then, add/edit HOME as the “variable name” with the intended location as the “variable value” (e.g., C:/Users/[user]/Documents, where you replace “user” with your HawkID).
Rprofile.site from the previous step
Sys.setenv("HOME" = "C:/Users/[specific user ID])/Documents").Rprofile file to the HOME directory and overwrite the original .Rprofile file (if it exists). You may have to show hidden files in order to see the file (PC: see Windows Explorer settings; Mac: Command+Shift+Dot)..Rprofile (not .Rprofile.Rprofile). Make sure there is a period at the beginning of the filename.RStudio. If the Rprofile.site and .Rprofile files are correctly set up, they should pre-populate your path location when you open R. If the contents of the Global Environment in RStudio are empty, your Rprofile.site and/or .Rprofile files are not set up correctly.
Error: could not find function "install.packages"), run the following line manually and then restart RStudio after the package finishes installing: install.packages("fortunes")
R/RStudio from saving your workspaces automatically using the following steps:
Tools → Global Options from the menus.Save workspace to .RData on exit to Never.OK.petersenlab R package using the following steps:
remotes package using the following command: install.packages("remotes")
petersenlab package using the following command: remotes::install_github("DevPsyLab/petersenlab")
ipconfig and press EnterIPv4 Address corresponds to your public IPEncrypt REDCap Token.R script: https://research-git.uiowa.edu/petersenlab/R-InitialSetup/blob/master/REDCap%20Credentials/Encrypt%20REDCap%20Token.R
REDCap Encryption Key.RData) was saved where you intended it to be saved on your local computer//lc-rs-store24.hpc.uiowa.edu/lss_itpetersen/Lab/Studies/School Readiness Study/Data Management/REDCap/Tokens/
REDCap Encryption Key.RData) to the comparable location of any other computers you own that you plan to access the data from
path variable you set in Rprofile.site) of every computer in order for it to be found by the Export Data.R script. The default location is: file.path(path, "GitHub/R/Data/REDCap Encryption Key.RData"), so if path is set as "C:/User/YourName", the file would be saved in: C:/User/YourName/GitHub/R/Data/REDCap Encryption Key.RData. The recommended location for GitHub repos is to create a folder titled GitHub in your Documents folder, and to put repos in the GitHub folder; it is NOT recommended to put git repos in a OneDrive folder because git files tend not to play nice with syncing services (archived at https://perma.cc/XZ6F-43G3; e.g., OneDrive, Dropbox)GitHub Desktop App (//lc-rs-store24.hpc.uiowa.edu/lss_itpetersen/Lab/Studies/School Readiness Study/Data Processing)
RStudio by using “Run as Administrator” (always open RStudio as an administrator so it has write access to the program files directory);Export Data.R script in R: https://research-git.uiowa.edu/petersenlab/srs/SRS-DataProcessing/blob/master/1.%20Export%20Data/Export%20Data.R \\lc-rs-store24.hpc.uiowa.edu\lss_itpetersen\Lab\Studies\School Readiness Study\Data Processing\1. Export Data\Export Data.R
RStudio, change the Graphics backend to Cairo: Tools → Global Options → Graphics
The petersenlab package is here: https://devpsylab.github.io/petersenlab. To install the petersenlab package, see instructions here.
To install and load R packages, see the instructions here.
To update packages, use the following code:
update.packages(checkBuilt = TRUE)One indication that the packages might not be updating to the latest version is seeing the same packages showing as needing an update after having run the update.packages() function. If this does not update the package(s) to the latest version, you may need to install the latest version of the package(s) from source (see the section on “Initial Set Up” of R for the software needed to install R packages from source):
update.packages(checkBuilt = TRUE, type = "source")R
Instructions adapted from: https://mirror.las.iastate.edu/CRAN/bin/windows/base/rw-FAQ.html#What_0027s-the-best-way-to-upgrade_003f (archived at https://perma.cc/W5QW-MA6Q)
R
R version into a directory that contains no spaces (see Step 2 in the Initial Set Up section above)Rprofile.site file, as described above, which installs packages to the common/shared “Packages” folder]:
R version folder, copy the current Rprofile.site file as a backup (Rprofile_BACKUP.site) and overwrite the original file with the lab’s version of Rprofile.site from here: https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/R%20Setup%20Files/Rprofile.site
R will run the file named Rprofile.site at initial runtime.R and RStudio to always run with administrator permissions.
R (C:\R\R-VERSION\bin\R.exe) and RStudio (C:\Program Files\RStudio\bin\RStudio.exe). Right-click it to open the contextual menu. Then, click or tap on “Properties”. In the Properties window, go to the Compatibility tab. At the bottom of the window, check the box next to the “Run this program as an administrator” option, and then click or tap on Apply or OK.R Compiler Tools for Rcpp on MacOS; see the instructions in the section on initial set up)R version and run update.packages(checkBuilt = TRUE, ask = FALSE), and install any necessary packagesRstudio Project
For each data analysis project (i.e., each GitLab/GitHub repo), create an RStudio Project. This helps keep your project files organized.
R Notebooks for “Computational Notebooks”Using R Notebooks for “Computational Notebooks” is helpful for reproducible code that can be shared with others. To create computational notebooks see the Markdown section on computational notebooks in the Data Analysis guides.
R scripts, use sections.
RStudio, use CTRL-Shift-R or “Code” - “Insert Section”R Notebooks/Markdown, use Headers and code chunks.
Ctrl+Alt+I; or click “Insert” button then “R”cbcl_ for the Child Behavior Checklist variablesprefix_thisIsTheVariableName
For reproducibility purposes, it is important not to save your workspace image (archived at https://perma.cc/9SCZ-L4DE). It is best practices to begin R each session with a clean workspace. If there is a .Rdata file in the same folder as the Rstudio Project, Rstudio will automatically load the objects into the workspace at the beginning of the session. This is problematic because those objects can interact/interfere with the code and can lead to problems with replicability for others who are running the code without those objects in the workspace. When you exit RStudio, RStudio asks if you want to “Save workspace image to [filepath]/.Rdata?” Make sure to select “Don’t Save”! However, do make sure to save your R scripts before exiting Rstudio.
R Markdown
https://www.spsanderson.com/steveondata/posts/2023-06-29/index.html (archived at https://perma.cc/9EXK-W99Y)
R scripts can be run automatically. For example, it can be helpful to have an R markdown report run automatically before the day begins.
Notepad app and create a file with the following syntax.
Location of R executable file\R CMD BATCH "Path location of script that should be automatically run"C:\R\4.1.3\bin\R CMD BATCH "R:\Lab\Studies\School Readiness Study\Data Processing\5. Reports\automatic_reports\Run_Reports_auto.R".bat file in the desired location.bat file has been created, search Task Scheduler in the search bar 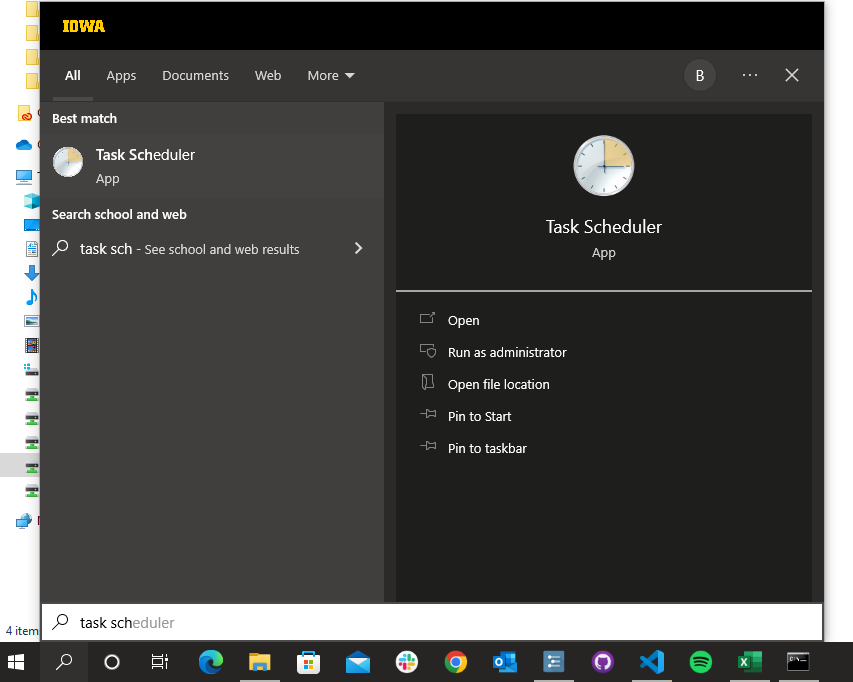
Actions selection bar, select Create basic Task...
Start a program
Program/script browse to the .bat file that was created in step 1 and select Next
Finish and the script is now configured to run automaticallyConditions of the task to ensure that the computer “wakes up” to execute the script
Conditions from the tabs across the top of the task windowWake the computer to run this task
R is updated, the path to the bin folder within R needs to be updated to reflect an accurate absolute path to R.
C:\R\4.1.3\bin\R CMD BATCH changed to C:\R\4.3.0\bin\R CMD BATCH
This error may appear if you are attempting to render a markdown file
pandoc version 1.12.3 or higher is required and was not found.The solution to this problem can be found at this link (archived at https://perma.cc/YX57-BPRS)
When using R to perform such actions as rendering sites and processing data, it can be useful to include code that commits and pushes relevant changes into the appropriate git repository. The steps below assume that you have Git and the GitHub desktop app installed. The instructions to download and configure git software can be found here
Install and/or load git2r package
install.packages("git2r")
library(git2r)Specify the path to your repository
repoPath <- "C:/Users/username/Documents/GitHub/exampleRepo"Open and define the repository
repo <- repository(repoPath)Stage changes to be committed
Stage all files in repo: r add(repo, "*")
Stage files with a specific file extension: r add(repo, "*.txt")
Stage files with one of any specified file extension: r add(repo, c("*.txt", "*.csv", "*.html"))
Stage files within a specific folder: r add(repo, "/folderName/*)
Stage one specific file: r add(repo, "fileName.txt)
Stage multiple specific files: r add(repo, c("thisFile.txt", "thatFile.txt", "anotherFile.txt"))
Commit the changes
Commit with a customized commit message: r commit(repo, "My commit message")
Commit with a generated message using system or environment variables:
Push changes
push(repo)Note: The code included above will prompt Git to authenticate your credentials by manually providing your username and password in a popup window. The push() function accepts a credentials = cred_user_pass("username", "password") argument that will “bypass” the need for user input by including the relevant credentials in the push() function call. One could provide their username and password directly in the script running this function, but other more-secure options include:
See here for an example of encryption and key creation
Using the encrypted credential and encryption key:
# Define key location
encryptionKeyLocation <- "path/to/key/location/myKey.RData"
# Load key into environment
load(encryptionKeyLocation)
# Read encrypted credential file using key
creds <- read.aes("path/to/encrypted/credentials/file.txt", key = key)
# Extract password from credential data
myPassword <- creds$password
# Push changes to git using credentials
push(repo, credentials = cred_user_pass("insertUsername", myPassword))try() Statementstry() statements can be very useful in cases where you would like code to continue running even if a certain line errors out. If the code inside of the statement results in an error, the try() statement will allow R to simply jump to the next line of code, rather than halting the entire process.
It is best practice to ensure that the smallest possible “unit” of code is wrapped in the try() statement rather than a large section. This is because the try() statement will treat everything within it as a “single” operation. Thus, an error ocurring early in the sequence within a try() statement will cause the remainder of the sequence to be skipped, and R will skip to the next line of code outside of the present try() statement.
For example:
The following code does NOT have try() statements around the smallest possible unit. If file_b.R were to error out, file_c.R would not run. In other words, file_c.R would be skipped because it occurs after file_b.R in the same try() statement.
On the other hand, the revised code below IS using try() statements around the smallest possible unit. If file_b.R were to error out, file_c.R would then begin to run. This is because each individual file being “sourced” is wrapped in its own try() statement.
A helpful post can be found here:
https://stackoverflow.com/questions/35852722/how-do-you-read-a-password-protected-excel-file-into-r (archived at https://perma.cc/U32Z-22VE)
install.packages("excel.link")
library("excel.link")
passwordProtectedBook <- xl.read.file(file.path("full path to workbook"), #Full path to workbook
password = "pass", #password
write.res.password="pass") #writing the reset passwordR
Occasionally, it can be helpful to send a Slack message using R. For example, if a script does not run, a Slack message can be sent to inform the appropriate team members. These instructions (archived at https://perma.cc/9CWJ-J5ZT) can largely be followed to set up R to send Slack messages. However, there are some differences:
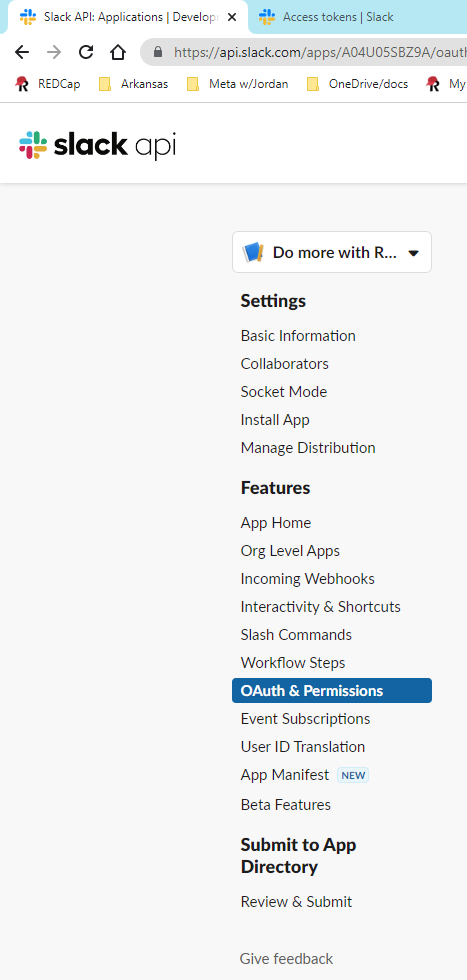
token category.
Oauth & Permissions
Once the configuration is complete, it is possible to send messages. For now, we have found it helpful to embed the slacks in the tryCatch function.
It is also possible to slack specific users with instructions found at this link (archived at https://perma.cc/59U5-V4GQ).
//n with a spaceMany notes in projects that are exported from REDCap come with spaces denotes as //n. Use the below code to make these fields more readable in the future.
gsub('\\n', ', ', df$notesField)R on a Network DriveWhen working with R on a network drive, it may be helpful to configure the project to store .Rproj.user on the local C:/ drive rather than on the network drive, which results in slow execution times.
For more info:
renv for Package Managementrenv is used for reproducibility, by helping with package management (tracking package versions, etc.):
https://rstudio.github.io/renv/articles/renv.html
Before updating a package locally, make sure that it is available in the Posit Package Manager (so it can be available to GitHub Actions):
https://packagemanager.posit.co
R:# 1. Update packages in package environment
renv::upgrade()
renv::update()
renv::snapshot()
# 2. Add/edit code
# 3. Update documentation
roxygen2::roxygenise()
# 4. Update package version (or can do this manually)
usethis::use_version()Then, build the package: Ctrl-Shift-B
Then, check the package: Ctrl-Shift-E
Then, run R CMD check
Then, install the package:
renv::install("C:/R/Packages/petersenlab") #PC
renv::install("/Library/Frameworks/R.framework/Packages/petersenlab") #MacTo install new packages in the package environment, run the following in R:
renv::install("NAME_OF_PACKAGE")or:
install.packages("NAME_OF_PACKAGE")R CMD check
.tar.gz file)
R console:devtools::build(pkg = "D:/Documents/GitHub/petersenlab")Open terminal in RStudio
.tar.gz file) is built, open a terminal window directly in RStudio by clicking on the “Terminal” tab at the bottom of RStudioRun R CMD check --as-cran
.tar.gz file) is located (commonly up one directory):Then, run R CMD check --as-cran followed by the name of your package tarball (making sure to update the package version in the filename). For example:
In the terminal:
Then, delete the created folder whose name begins with “.Rd2pdf…”
In the R Console, build the source package in R (or using the instructions described above):
devtools::build(pkg = "D:/Documents/GitHub/petersenlab")In the terminal (making sure to update the package version in the filename):
no visible binding for global variable; Undefined global functions or variables
For example:
Solution: set each variable to NULL in the package function before it is mentioned. For example:
predictorVal_centered <- moderatorVal_centered <- NULLR CMD check via GitHub Actionsusethis::use_github_action("check-standard")Install Package: ‘Ctrl + Shift + B’
Check Package: ‘Ctrl + Shift + E’
Test Package: ‘Ctrl + Shift + T’
renv::install("C:/R/Packages/petersenlab")
renv::snapshot()
renv::install()pkgdown
Run once to configure your package to use pkgdown:
usethis::use_pkgdown()Then use pkgdown to build your website:
pkgdown::build_site().R file with the function_pkgdown.yml filerenv::upgrade()renv::update()renv::snapshot()roxygen2::roxygenise()R CMD checkdevtools::build(pkg = "D:/Documents/GitHub/petersenlab/inst/extdata/testpackage1")
devtools::build(pkg = "D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2")
install.packages("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage1_0.1.0.tar.gz", repos = NULL, source = TRUE)
install.packages("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2_0.1.0.tar.gz", repos = NULL, source = TRUE)
remotes::install_local("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2_0.1.0.tar.gz")
remotes::install_local("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2_0.1.0.tar.gz")Official documentation for CRAN:
Unofficial documentation:
To troubleshoot R issues, be resourceful. Try googling the error message or issue you are experiencing. See here for a list of places you can pose R-related questions for help. In addition, particular errors/warnings/issues are included below:
[object name] not found)
CTRL-F to search for it is a good way to check these thingsnon-numeric argument to binary operator indicate that the function you are trying to use is expecting to recieve numeric data as an input but one or more of the inputs are not numeric
class(data$variable) (replace “data” and “variable” with whatever you are trying to invesitgate) is useful for determining how R is interpreting your data. Often, data that look like numbers can end up being stored as a character (text) variable. This can be due to import/export processes or due to an issue with data entry.
00 is entered instead of 0 in a numeric field R is likely to interpret the variable as a character instead of a number when importing the datavariable.x or variable.y)variable.x instead of variable) or deal with the duplicated columns before/during the joining process (e.g., include duplicate columns in the by argument of joining function or remove the redundant column from one of the datasets to be joined)NA (missing) or NaN (not a number) values. Such values may not be an issue or error in all cases, but if you are not expecting that as a result of your computations, this might suggest an issue with the code and/or data structureCode can be wrapped in try() statements so that if an error occurs, the script will simply “skip” to the next line of code. These try() statements are particularly useful when rendering a site; if they are not used, a single error will halt the entire process. try() statements allow the code to continue running in spite of a line or two erroring out.
If you have already incorporated try() statements and are still experiencing fatal errors in code, check to make sure that the smallest possible “unit” of code is wrapped in try() statements, rather than large sections of code. See here for more information.
PACKAGENAME package in FILEPATH library will not be updatedThis warning might indicate that it cannot update the package because it is part of the base R installation. To fix this:
R installation in the library directory of the R installation (e.g., C:\R\R-4.3.1\library\PACKAGENAME\)C:\R\Packages\PACKAGENAME\); if it is, delete it from there as wellR and run install.packages("PACKAGENAME") to reinstall the packageR
R installation, move the installed package folder C:\R\Packages\PACKAGENAME\ to the library directory of the R installation (e.g., C:\R\R-4.3.1\library\PACKAGENAME\)R version 4.5.2 (2025-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
time zone: UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] htmlwidgets_1.6.4 compiler_4.5.2 fastmap_1.2.0 cli_3.6.5
[5] tools_4.5.2 htmltools_0.5.9 otel_0.2.0 yaml_2.3.12
[9] rmarkdown_2.30 knitr_1.51 jsonlite_2.0.0 xfun_0.56
[13] digest_0.6.39 rlang_1.1.7 evaluate_1.0.5 ---
title: "R"
---
> "*`R` will always be arcane to those who do not make a serious effort to learn it. It is not meant to be intuitive and easy for casual users to just plunge into. It is far too complex and powerful for that. But the rewards are great for serious data analysts who put in the effort.*"
— [Berton Gunter R-help August 2007](https://www.brodrigues.co/blog/2022-06-02-arcane/) (archived at <https://perma.cc/KY9N-2FTT>)
> "Evelyn Hall: I would like to know how (if) I can extract some of the information from the summary of my nlme.
>
> Simon Blomberg: This is `R`. There is no if. Only how."
— [Evelyn Hall and Simon 'Yoda' Blomberg, R-help April 2005](https://www.brodrigues.co/blog/2022-06-02-arcane/) (archived at <https://perma.cc/KY9N-2FTT>)
# Learning `R` {#sec-learn}
## Base R
Here are a slew of resources for learning base `R` (in addition to the documents in the lab's [Primer Articles folder](https://drive.google.com/drive/u/0/folders/1U0pNrjF2_tIPU6BiyCv0HqbOUA5P516r)):
- `R for Data Science` book: <https://r4ds.hadley.nz>
- Tutorials from the book are available via the `r4ds.tutorials` package: <https://ppbds.github.io/r4ds.tutorials/>
- Use this Intro to `R`: <https://www.statmethods.net> (archived at <https://perma.cc/SPQ2-DJKM>)
- I used this resource considerably when I was learning `R`
- `Codeacademy` has free online courses:
- Basics of using R: <https://www.codecademy.com/learn/learn-r> (archived at <https://perma.cc/JHS2-EYUU>)
- Learn how to become a better coder: <https://www.r-bloggers.com/10-top-tips-for-becoming-a-better-coder/> (archived at <https://perma.cc/H3W7-QDAM>)
- How to become fluent in `R`: <https://www.sharpsightlabs.com/blog/are-you-fluent-r/> (archived at <https://perma.cc/RWL5-GTWV>)
- Video training courses in `R` skills: <https://www.pluralsight.com/search?q=R> (archived at <https://perma.cc/U4ZZ-UE4X>)
- Browse the `Cookbook for R` to find solutions to common tasks and problems: <http://www.cookbook-r.com> (archived at <https://perma.cc/5ERJ-VRFR>)
- Browse the `R Graph Gallery` to find examples of various graphs: <https://r-graph-gallery.com>
- Free `Coursera` courses on `R`: <https://pairach.com/2012/12/22/learn-to-use-r-for-free-with-coursera/> (archived at <https://perma.cc/XJ5U-5E4W>)
- MOOCs and courses to learn `R`: <https://www.r-bloggers.com/moocs-and-courses-to-learn-r/> (archived at <https://perma.cc/MQ25-E3ME>)
- Watch these videos from `Coursera`: <https://blog.revolutionanalytics.com/2012/12/coursera-videos.html> (archived at <https://perma.cc/6FU4-PAQW>)
- `RStudio` Webinars: <https://www.rstudio.com/resources/webinars/> (archived at <https://perma.cc/6RNR-98JB>)
- UCLA Stats Website: <https://stats.idre.ucla.edu/r/> (archived at <https://perma.cc/M3N6-96RY>)
- Take this Introduction to `R` course: <https://www.datacamp.com/courses/free-introduction-to-r> (archived at <https://perma.cc/6ZM9-37L9>)
- Teaching `R` in a Kinder, Gentler, More Effective Manner: <https://github.com/matloff/TidyverseSkeptic> (archived at <https://perma.cc/QU9F-FBS8>)
- Learn `R` interactively with `swirl`: <https://swirlstats.com> (archived at <https://perma.cc/TT9V-U663>)
- Use the `learnr` package: <https://blog.rstudio.com/2017/07/11/introducing-learnr/> (archived at <https://perma.cc/XGJ8-EXSN>)
- You will sometimes find relevant articles on `R-bloggers`: <https://www.r-bloggers.com> (archived at <https://perma.cc/EL3X-ZXBB>)
## Statistics (using `R`)
The following are resources for learning statistics using `R`.
- `R` for Data Science: <https://r4ds.hadley.nz>
- UCLA Stats Website: <https://stats.oarc.ucla.edu/other/dae/> (archived at <https://perma.cc/FVG5-5X82>)
- `Codeacademy` has free online courses:
- Descriptive analysis and basic hypothesis testing: <https://www.codecademy.com/learn/learn-statistics-with-r>
- Free textbook on Learning Statistics with `R`: <https://learningstatisticswithr.com> (archived at <https://perma.cc/SY87-5GCX>)
- An excellent introductory textbook on Discovering Statistics using `R`: <https://www.amazon.com/Discovering-Statistics-Using-Andy-Field/dp/1446200469> (archived at <https://perma.cc/9LGU-7ZME>)
- My textbook that teaches `R`/statistics through the lens of fantasy football: <https://isaactpetersen.github.io/Fantasy-Football-Analytics-Textbook>
## `tidyverse`
The following are resources for learning `tidyverse`, which is a collection of `R` packages for data management:
- <https://www.tidyverse.org/learn/> (archived at <https://perma.cc/5ZUM-XGES>)
- <https://www.linkedin.com/learning/learning-the-r-tidyverse/welcome?u=42459020> (archived at <https://perma.cc/TD56-FX8R>)
## Getting Help/Questions {#sec-questions}
If you have `R` questions, you can ask them in a number of places:
- Forums:
- `Posit`: <https://forum.posit.co>
- `StackOverflow`: <https://stackoverflow.com/questions/tagged/r>
- `Reddit`: <https://www.reddit.com/r/rstats/>
- The `R` mailing list: <https://stat.ethz.ch/mailman/listinfo/r-help>
The following article provides additional resources and good guidance: <https://www.r-bloggers.com/where-to-get-help-with-your-r-question/> (archived at <https://perma.cc/4RRE-KL33>).
When posting a question on forums or mailing lists, keep a few things in mind:
- Read the posting guidelines before posting!
- Be respectful of other people and their time.
`R` is free software.
People are offering their free time to help.
They are under no obligation to help you.
If you are disrespectful or act like they owe you anything, you will rub people the wrong way and will be less likely to get help.
- Provide a minimal, reproducible example.
Providing a minimal, reproducible example can be crucial for getting a helpful response.
By going to the trouble of creating a minimal, reproducible example and identifying the minimum conditions necessary to reproduce the issue, you will often figure out how to resolve it.
Here are guidelines on providing a minimal, reproducible example: <https://stackoverflow.com/help/minimal-reproducible-example> (archived at <https://perma.cc/6NUB-UTYF>).
Here are a good example and guidelines for providing a minimal, reproducible example in `R`: <https://stackoverflow.com/a/5963610> (archived at <https://perma.cc/PC9L-DQZG>).
Provide a `reprex` whenever possible: <https://reprex.tidyverse.org>.
# Initial Set Up {#sec-setup}
Note: many of these initial setup steps described below are not necessary for general use; many of these steps are necessary only for using lab-related repositories (e.g., to gain API access to export data from `REDCap`, to use absolute paths rather than relative paths so repos can communicate with each other, etc.).
1. Make sure you are logged onto a computer that can access the lab server (either a lab computer, or a computer you can VPN into the lab server), and that you have admin access to install and uninstall software
1. Install `R` (<https://www.r-project.org>) into a directory that contains no spaces; On PC, change the location from the default `C:/Program Files/R/[R-VERSION]` (which contains a space) to `C:/R/[R-VERSION]` ([which does not contain any spaces](https://cran.r-project.org/doc/manuals/r-release/R-admin.html#Installing-R-under-Windows); archived at <https://perma.cc/6VMX-3LYX>—this is because [some packages that require compilation to install cannot read filepaths with spaces](https://cran.r-project.org/doc/manuals/r-release/R-admin.html#Installation); archived at <https://perma.cc/XA3V-JTPY>); may have to right click and "Run As Administrator"
- If `R` was already installed in a directory that contains spaces (e.g., `C:/Program Files/R/[R-VERSION]`), uninstall `R` before installing it in a directory that doesn't contain spaces
1. Install `RStudio` Desktop (<https://www.rstudio.com/products/rstudio/download/>) in the main program files directory; may have to right click and "Run As Administrator".
`RStudio` is the best available graphical user interface for R.
1. Set the executables for `R` and `RStudio` to always run with administrator permissions.
- If on Windows, open File Explorer and find the main executable of `R` (`C:/R/[R-VERSION]/bin/R.exe`) and `RStudio` (`C:/Program Files/RStudio/bin/RStudio.exe`).
Right-click it to open the contextual menu.
Then, click or tap on "Properties".
In the Properties window, go to the Compatibility tab.
At the bottom of the window, check the box next to the "Run this program as an administrator" option, and then click or tap on Apply or OK.
1. Install tools to allow you to compile `R` packages so you can install packages from source, if necessary (i.e., if package binaries are not available):
- If on Windows, install [Rtools](https://cran.r-project.org/bin/windows/Rtools/); may have to right click and "Run As Administrator"
- If on Mac, install [R Compiler Tools for Rcpp on MacOS](https://thecoatlessprofessor.com/programming/cpp/r-compiler-tools-for-rcpp-on-macos/) (archived at <https://perma.cc/B35J-R22X>)
1. Set up `git`, `GitLab`, and the `GitHub Desktop` App in the main program files directory; may have to right click and "Run As Administrator"; For instructions setting up and using `GitLab`, see here: <https://devpsylab.github.io/DataAnalysis/git.html#toBegin>
1. The `Rprofile.site` file in the `etc` folder of the `R` installation directory is the code that is run for ***every user*** at the beginning each time you load R. We will update the default `Rprofile.site` file with the lab's `Rprofile.site` file so `R` installs packages in the correct location, sets the default package repository, updates packages, and gives you a fortune cookie.
To do this, perform the following steps:
- Rename the `Rprofile.site` file in the `R` installation directory (`C:/R/R-[InsertVersionNumber]/etc/Rprofile.site`) to be `Rprofile_BACKUP.site`
- Download the lab's `Rprofile.site` file located in this repository at the following location (<https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/R%20Setup%20Files/Rprofile.site>), and paste it into the `R` installation directory (PC: `C:/R/R-[InsertVersionNumber]/etc/Rprofile.site`; Mac: `/Library/Frameworks/R.framework/Versions/[InsertVersionNumber]/Resources/etc/Rprofile.site`)
1. The `.Rprofile` file in the user's `Documents` folder is the code that is run for ***the particular user*** at the beginning each time you load R.
We will update the default `.Rprofile` file (if there is one) with the lab's `.Rprofile` file so `R` knows which computer you are using and which path to use (relative to where your `R` projects are located).
To do this, perform the following steps:
- Download the lab's `.Rprofile` template file in this repository at the following location, and make sure to remove anything besides `.Rprofile` in the filename: <https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/R%20Setup%20Files/.Rprofile>
- Open the lab's `.Rprofile` file, and revise it with your HawkID
- Revise the lab's `.Rprofile` file with the local path to the `Documents` folder for each of the computers you will use to access `R` (e.g., home computer, work computer, laptop).
Make sure to use forward slashes (`/`), not back slashes (`\`) in the path.
- You will save the file in your `HOME` directory.
To find the `HOME` directory, open `R` and type the following command: `Sys.getenv("HOME")`—the output of the command is the location of your `HOME` directory; If this is a lab computer, it may be located here: `//home.iowa.uiowa.edu/[user]/Documents`.
If this is your personal computer, it may be located here: PC: `C:/Users/[user]/Documents`; Mac: `/Users/[user]`.
Then close R.
- If your `HOME` directory is in a OneDrive folder (or another cloud-based sync folder), you will want to change the directory of your `HOME` path so that it is not in a OneDrive folder.
To do that, open [`Environment Variables`](https://superuser.com/questions/949560/how-do-i-set-system-environment-variables-in-windows-10) (archived at <https://perma.cc/A2E5-B5VA>) in Windows.
Then, add/edit `HOME` as the "variable name" with the intended location as the "variable value" (e.g., `C:/Users/[user]/Documents`, where you replace "user" with your HawkID).
- You may also solve this issue by placing the following command in the `Rprofile.site` from the previous step
- `Sys.setenv("HOME" = "C:/Users/[specific user ID])/Documents")`
- Move the revised `.Rprofile` file to the `HOME` directory and overwrite the original `.Rprofile` file (if it exists).
You may have to show hidden files in order to see the file (PC: see Windows Explorer settings; Mac: Command+Shift+Dot).
- Make sure to show filename extensions in your file explorer window, and make sure the file is named `.Rprofile` (not `.Rprofile.Rprofile`).
Make sure there is a period at the beginning of the filename.
1. Run `RStudio`.
If the `Rprofile.site` and `.Rprofile` files are correctly set up, they should pre-populate your `path` location when you open R.
If the contents of the `Global Environment` in `RStudio` are empty, your `Rprofile.site` and/or `.Rprofile` files are not set up correctly.
- If you get this error (`Error: could not find function "install.packages"`), run the following line manually and then restart `RStudio` after the package finishes installing: `install.packages("fortunes")`
1. For [reproducibility purposes](#sec-dontSaveWorkspace), prevent `R`/`RStudio` from saving your workspaces automatically using the following steps:
- With RStudio running, choose `Tools → Global Options` from the menus.
- In the Options dialog, change the value for `Save workspace to .RData on exit` to `Never`.
- Click `OK`.
1. Install the `petersenlab` `R` package using the following steps:
- Install the `remotes` package using the following command: `install.packages("remotes")`
- Install the `petersenlab` package using the following command: `remotes::install_github("DevPsyLab/petersenlab")`
1. Request an [API token](https://redcap.icts.uiowa.edu/redcap/api/help/?content=tokens) for the following REDCap project(s); note: please check with Dr. P before requesting an API token.
In general, RAs should not have an API token.
- [School Readiness Study](https://redcap.icts.uiowa.edu/redcap/redcap_v12.4.1/index.php?pid=4941)
- [School Readiness Study Screening](https://redcap.icts.uiowa.edu/redcap/redcap_v12.4.1/index.php?pid=4958)
- [201701837 - Mechanisms in the Development of Self-Regulation, School Readiness, and Behavior Problems [Data Extraction]](https://redcap.icts.uiowa.edu/redcap/redcap_v12.4.1/index.php?pid=11233)
- [School Readiness Study - Prospective Participants](https://redcap.icts.uiowa.edu/redcap/redcap_v12.4.1/index.php?pid=10440)
1. Complete the survey(s) provided by REDCap administration
- The first section will be some basic questions about the planned usage of the API(s)
- **You will need to provide the public IP address for your workspace computer**
- To obtain the public IP address for a Windows PC:
- Open Command Prompt
- Type in `ipconfig` and press Enter
- The `IPv4 Address` corresponds to your public IP
1. When your API token has been approved for these projects, open the `Encrypt REDCap Token.R` script:
<https://research-git.uiowa.edu/petersenlab/R-InitialSetup/blob/master/REDCap%20Credentials/Encrypt%20REDCap%20Token.R>
1. Revise the API tokens to reflect yours, then run the script to save your encrypted credentials on the lab server and your encryption key on your local computer
- Verify that the Encryption Key (`REDCap Encryption Key.RData`) was saved where you intended it to be saved on your local computer
- Verify that a file named with your HawkID was saved here: `//lc-rs-store24.hpc.uiowa.edu/lss_itpetersen/Lab/Studies/School Readiness Study/Data Management/REDCap/Tokens/`
1. Copy the Encryption Key (`REDCap Encryption Key.RData`) to the comparable location of any other computers you own that you plan to access the data from
- The file has to be in the comparable location (relative to the `path` variable you set in `Rprofile.site`) of every computer in order for it to be found by the `Export Data.R` script.
The default location is: `file.path(path, "GitHub/R/Data/REDCap Encryption Key.RData")`, so if `path` is set as `"C:/User/YourName"`, the file would be saved in: `C:/User/YourName/GitHub/R/Data/REDCap Encryption Key.RData`.
The recommended location for `GitHub` repos is to create a folder titled `GitHub` in your `Documents` folder, and to put repos in the `GitHub` folder; it is *NOT* recommended to put `git` repos in a OneDrive folder because [`git` files tend not to play nice with syncing services](https://stackoverflow.com/questions/19305033/why-is-putting-git-repositories-inside-of-a-dropbox-folder-not-recommended) (archived at <https://perma.cc/XZ6F-43G3>; e.g., OneDrive, Dropbox)
1. Add the SRS Data Processing repo from the lab drive to your `GitHub Desktop` App (`//lc-rs-store24.hpc.uiowa.edu/lss_itpetersen/Lab/Studies/School Readiness Study/Data Processing`)
- For instructions, see the section on "[How to add a pre-existing repo from the lab drive (RDSS/network share) to your computer](https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/GitLab/GitLab%20Instructions.md#how-to-add-a-pre-existing-repo-from-the-lab-drive-rdssnetwork-share-to-your-computer)" here: <https://research-git.uiowa.edu/petersenlab/R-InitialSetup/blob/master/GitLab/GitLab%20Instructions.md>
1. Open `RStudio` by using "Run as Administrator" (always open `RStudio` as an administrator so it has write access to the program files directory);
1. Open the `Export Data.R` script in R:
<https://research-git.uiowa.edu/petersenlab/srs/SRS-DataProcessing/blob/master/1.%20Export%20Data/Export%20Data.R>
`\\lc-rs-store24.hpc.uiowa.edu\lss_itpetersen\Lab\Studies\School Readiness Study\Data Processing\1. Export Data\Export Data.R`
1. Ensure your HawkID and location of your encryption key in the script are correct, and then run the script to verify that you can export data from REDCap
1. For antialiased plots in `RStudio`, change the Graphics backend to `Cairo`:
`Tools → Global Options → Graphics`
# Lab Package {#sec-petersenlab}
The [`petersenlab` package](https://devpsylab.github.io/petersenlab) is here: <https://devpsylab.github.io/petersenlab>.
To install the [`petersenlab` package](https://devpsylab.github.io/petersenlab), see instructions [here](dataManagement.qmd#sec-labFunctions).
# Install Packages {#sec-installPackages}
To install and load `R` packages, see the instructions [here](dataManagement.qmd#sec-loadInstallPackages).
# Update Packages {#sec-updatePackages}
To update packages, use the following code:
```{r}
#| eval: false
update.packages(checkBuilt = TRUE)
```
One indication that the packages might not be updating to the latest version is seeing the same packages showing as needing an update after having run the `update.packages()` function.
If this does not update the package(s) to the latest version, you may need to install the latest version of the package(s) from source (see the section on "[Initial Set Up](#sec-setup)" of `R` for the software needed to install R packages from source):
```{r}
#| eval: false
update.packages(checkBuilt = TRUE, type = "source")
```
# Update `R` {#sec-updateR}
Instructions adapted from: <https://mirror.las.iastate.edu/CRAN/bin/windows/base/rw-FAQ.html#What_0027s-the-best-way-to-upgrade_003f> (archived at <https://perma.cc/W5QW-MA6Q>)
1. Uninstall `R`
1. Install the new `R` version into a directory that contains no spaces (see Step 2 in the [Initial Set Up](#sec-setup) section above)
1. [You only need to do this step if you installed packages in the R-version-specific "Library" folder rather than the common/shared "Packages" folder—that is, you don't need to do this step if you used the lab's `Rprofile.site` file, as described above, which installs packages to the common/shared "Packages" folder]:
- Copy installed packages in the "Library" folder to the "Library" folder in the new installation
1. In new `R` version folder, copy the current `Rprofile.site` file as a backup (`Rprofile_BACKUP.site`) and overwrite the original file with the lab's version of `Rprofile.site` from here: <https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/R%20Setup%20Files/Rprofile.site>
- `R` will run the file named `Rprofile.site` at initial runtime.
1. Set the executables for `R` and `RStudio` to always run with administrator permissions.
- If on Windows, open File Explorer and find the main executable of `R` (`C:\R\R-VERSION\bin\R.exe`) and `RStudio` (`C:\Program Files\RStudio\bin\RStudio.exe`).
Right-click it to open the contextual menu.
Then, click or tap on "Properties".
In the Properties window, go to the Compatibility tab.
At the bottom of the window, check the box next to the "Run this program as an administrator" option, and then click or tap on Apply or OK.
1. Make sure you have the latest version of the tools necessary to compile packages from source (i.e., Rtools for Windows or `R` Compiler Tools for Rcpp on MacOS; see the instructions in the section on [initial set up](#sec-setup))
1. Open the new `R` version and run `update.packages(checkBuilt = TRUE, ask = FALSE)`, and install any necessary packages
1. Close R
1. Delete anything left of the old installation
# Style Guide and Best Practices {#sec-bestPractices}
## Create `Rstudio Project`
For each data analysis project (i.e., each [`GitLab`/`GitHub`](#sec-git) repo), create an RStudio Project.
This helps keep your project files organized.
## Use `R` Notebooks for "Computational Notebooks"
Using `R` Notebooks for "Computational Notebooks" is helpful for reproducible code that can be shared with others.
To create computational notebooks see the `Markdown` section on [computational notebooks](markdown.qmd#sec-computationalNotebook) in the Data Analysis guides.
## Separate sections in code
- In `R` scripts, use sections.
- To insert a section in `RStudio`, use `CTRL-Shift-R` or "Code" - "Insert Section"
- In `R` Notebooks/Markdown, use Headers and code chunks.
- Headers: 1, 2, or 3 pound signs
- Code Chunks: `Ctrl+Alt+I`; or click "Insert" button then "R"
## Naming variables
- Use meaningful variable names; we want to know what a variable represents without having to consult an external codebook for every variable
- Variable names should include the prefix for the measure followed by an underscore
- e.g., `cbcl_` for the Child Behavior Checklist variables
- Use lower camel case for variable naming
- e.g., `prefix_thisIsTheVariableName`
- Do **not** include spaces in variable names
## Comment code frequently and clearly!
It is important to comment code frequently and clearly.
You want you (and others) to easily be able to understand your code if you come back to it several years later!
## Don't save your workspace image {#sec-dontSaveWorkspace}
For reproducibility purposes, it is important [**not** to save your workspace image](https://www.r-bloggers.com/2017/04/using-r-dont-save-your-workspace/) (archived at <https://perma.cc/9SCZ-L4DE>).
It is best practices to begin `R` each session with a clean workspace.
If there is a `.Rdata` file in the same folder as the `Rstudio Project`, Rstudio will automatically load the objects into the workspace at the beginning of the session.
This is problematic because those objects can interact/interfere with the code and can lead to problems with replicability for others who are running the code without those objects in the workspace.
When you exit `RStudio`, `RStudio` asks if you want to "Save workspace image to `[filepath]/.Rdata`?"
Make sure to select "Don't Save"!
However, do make sure to save your `R` scripts before exiting Rstudio.
# Data Management
- The lab's [Data Analysis Guides on Data Management](dataManagement.qmd)
- Tidyverse: <https://www.tidyverse.org>
- University of Iowa Workshops
# Saving Plots
`png(); dev.off()`
- The lab's [Data Analysis Guides on Figures](figures.qmd)
# Saving Output
`R Markdown`
- The lab's [Data Analysis Guides on Markdown](markdown.qmd)
- <https://rmarkdown.rstudio.com/lesson-1.html> (archived at <https://perma.cc/8SQH-N68X>)
- <https://www.rstudio.com/wp-content/uploads/2015/02/rmarkdown-cheatsheet.pdf> (archived at <https://perma.cc/3NMT-4L25>)
- <https://bookdown.org/yihui/rmarkdown/> (archived at <https://perma.cc/UJC8-ZZVC>)
# Shortcuts
- Run selected line(s) of code: Ctrl + Enter
- Comment/uncomment code: Ctrl + Shift + C
- Pipe: Ctrl + Shift + M
- Insert Code Chunk: Ctrl + Alt + I
- Assignment operator: Alt + - (alt-dash)
- Select multiple lines: Ctrl + Alt, up or down; or Alt + drag mouse
- Search: Ctrl + Shift + F
- Show all keyboard shortcuts: Alt + Shift + K
# Statistics Examples
- [Bayesian Analysis](bayesian.qmd)
- [Data Management](dataManagement.qmd)
- [Developmental Scaling](developmentalScaling.qmd)
- [Exploratory Data Analysis](eda.qmd)
- [Factor Analysis](factorAnalysis.qmd)
- [Hierarchical Linear Modeling](hlm.qmd)
- [Item Response Theory](irt.qmd)
- [Longitudinal Data Analysis](lda.qmd)
- [Mediation](sem.qmd#sec-mediation)
- [Moderation/Interaction](regression.qmd#sec-moderation)
- [Multiple Imputation](multipleImputation.qmd)
- [Principal Component Analysis](pca.qmd)
- [Regression](regression.qmd)
- [Structural Equation Modeling](sem.qmd)
# Running Scripts Automatically with Windows
<https://www.spsanderson.com/steveondata/posts/2023-06-29/index.html> (archived at <https://perma.cc/9EXK-W99Y>)
`R` scripts can be run automatically.
For example, it can be helpful to have an `R` markdown report run automatically before the day begins.
1. Open the `Notepad` app and create a file with the following syntax.
- `Location of R executable file\R CMD BATCH "Path location of script that should be automatically run"`
- **Example:**
- `C:\R\4.1.3\bin\R CMD BATCH "R:\Lab\Studies\School Readiness Study\Data Processing\5. Reports\automatic_reports\Run_Reports_auto.R"`
1. Save the file as a `.bat` file in the desired location
1. Once the `.bat` file has been created, search `Task Scheduler` in the search bar

1. In the `Actions` selection bar, select `Create basic Task...`
1. Name the task and provide a description
1. Next, set the trigger for the new task (i.e., how often the task should run)
1. Set the action for the task by selecting `Start a program`
1. Under, `Program/script` browse to the `.bat` file that was created in step 1 and select `Next`
1. Click `Finish` and the script is now configured to run automatically
1. If you wish to schedule a task to run during a time at which your computer is likely to be asleep, you'll need to change the `Conditions` of the task to ensure that the computer "wakes up" to execute the script
- In the Task Scheduler Library, select the task you just created
- The task should appear on the lower half of the screen (double click to open in a new window).
Select `Conditions` from the tabs across the top of the task window
- Select `Wake the computer to run this task`
- Note that elevated account permissions may be required to set this condition
1. **Note: When `R` is updated, the path to the `bin` folder within `R` needs to be updated to reflect an accurate absolute path to R.**
- Example: `C:\R\4.1.3\bin\R CMD BATCH` changed to `C:\R\4.3.0\bin\R CMD BATCH`
## Troubleshooting
### Pandoc error
This error may appear if you are attempting to render a markdown file
```
pandoc version 1.12.3 or higher is required and was not found.
```
The solution to this problem [can be found at this link](https://stackoverflow.com/questions/28432607/pandoc-version-1-12-3-or-higher-is-required-and-was-not-found-r-shiny) (archived at <https://perma.cc/YX57-BPRS>)
# Using R Script to Commit and Push Changes {#sec-gitPush}
When using R to perform such actions as rendering sites and processing data, it can be useful to include code that commits and pushes relevant changes into the appropriate `git` repository.
The steps below assume that you have `Git` and the `GitHub` desktop app installed.
The instructions to download and configure `git` software can be found [here](git.Rmd#sec-toBegin)
1. Install and/or load `git2r` package
```r
install.packages("git2r")
library(git2r)
```
1. Specify the path to your repository
```r
repoPath <- "C:/Users/username/Documents/GitHub/exampleRepo"
```
1. Open and define the repository
```r
repo <- repository(repoPath)
```
1. Stage changes to be committed
- Stage all files in repo:
```r
add(repo, "*")
```
- Stage files with a specific file extension:
```r
add(repo, "*.txt")
```
- Stage files with one of any specified file extension:
```r
add(repo, c("*.txt", "*.csv", "*.html"))
```
- Stage files within a specific folder:
```r
add(repo, "/folderName/*)
```
- Stage one specific file:
```r
add(repo, "fileName.txt)
```
- Stage multiple specific files:
```r
add(repo, c("thisFile.txt", "thatFile.txt", "anotherFile.txt"))
```
1. Commit the changes
- Commit with a customized commit message:
```r
commit(repo, "My commit message")
```
- Commit with a generated message using system or environment variables:
```r
# example: include system date in commit message
date <- format(Sys.Date(), format = %Y-%m-%d)
message <- paste(date, "daily site update", sep = " ")
commit(repo, message)
```
1. Push changes
```r
push(repo)
```
**Note:** The code included above will prompt `Git` to authenticate your credentials by manually providing your username and password in a popup window.
The `push()` function accepts a `credentials = cred_user_pass("username", "password")` argument that will "bypass" the need for user input by including the relevant credentials in the `push()` function call.
One could provide their username and password directly in the script running this function, but other more-secure options include:
- Using a [Personal Access Token](https://docs.gitlab.com/user/profile/personal_access_tokens/)
- Using an [SSH key](https://git-scm.com/book/en/v2/Git-on-the-Server-Generating-Your-SSH-Public-Key)
- Encrypting credential information and storing the encrypted credential file somewhere accessible for use
- See [here](https://research-git.uiowa.edu/PetersenLab/R-InitialSetup/-/blob/master/REDCap%20Credentials/Encrypt%20REDCap%20Token.R) for an example of encryption and key creation
- Using the encrypted credential and encryption key:
```r
# Define key location
encryptionKeyLocation <- "path/to/key/location/myKey.RData"
# Load key into environment
load(encryptionKeyLocation)
# Read encrypted credential file using key
creds <- read.aes("path/to/encrypted/credentials/file.txt", key = key)
# Extract password from credential data
myPassword <- creds$password
# Push changes to git using credentials
push(repo, credentials = cred_user_pass("insertUsername", myPassword))
```
# Using `try()` Statements {#sec-try}
[`try()` statements](https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/try) can be very useful in cases where you would like code to continue running even if a certain line errors out.
If the code inside of the statement results in an error, the `try()` statement will allow R to simply jump to the next line of code, rather than halting the entire process.
It is best practice to ensure that the smallest possible "unit" of code is wrapped in the `try()` statement rather than a large section.
This is because the `try()` statement will treat everything within it as a "single" operation.
Thus, an error ocurring early in the sequence within a `try()` statement will cause the remainder of the sequence to be skipped, and R will skip to the next line of code outside of the present `try()` statement.
For example:
The following code does NOT have `try()` statements around the smallest possible unit.
If `file_b.R` were to error out, `file_c.R` would not run.
In other words, `file_c.R` would be skipped because it occurs after `file_b.R` in the **same** `try()` statement.
```r
try({
source(path/to/file/file_a.R)
source(path/to/file/file_b.R)
source(path/to/file_c.R)
})
```
On the other hand, the revised code below IS using `try()` statements around the smallest possible unit.
If `file_b.R` were to error out, `file_c.R` would then begin to run.
This is because each individual file being "sourced" is wrapped in its **own** `try()` statement.
```r
try({source(path/to/file/file_a.R)})
try({source(path/to/file/file_b.R)})
try({source(path/to/file_c.R)})
```
# Reading Password Protected Excel Databases
A helpful post can be found here: <br>
<https://stackoverflow.com/questions/35852722/how-do-you-read-a-password-protected-excel-file-into-r> (archived at <https://perma.cc/U32Z-22VE>)
```{r}
#| eval: false
install.packages("excel.link")
library("excel.link")
passwordProtectedBook <- xl.read.file(file.path("full path to workbook"), #Full path to workbook
password = "pass", #password
write.res.password="pass") #writing the reset password
```
# Sending slacks with `R`
Occasionally, it can be helpful to send a Slack message using `R`.
For example, if a script does not run, a Slack message can be sent to inform the appropriate team members.
[These instructions](https://www.infoworld.com/article/3402657/how-to-slack-from-r.html) (archived at <https://perma.cc/9CWJ-J5ZT>) can largely be followed to set up `R` to send Slack messages.
However, there are some differences:
1. When setting up the configuration file, use the below template.
The slack API token should be placed in the `token` category.
- Note the token will need to be updated every 30 days.
You can generate a new token by navigating to the [Slack API](https://api.slack.com/apps) and selecting `Oauth & Permissions`
- 
```r
token: YOUR_FULL_API_TOKEN
channel: #general
username: slackr
incoming_webhook_url: https://hooks.slack.com/services/XXXXX/XXXXX/XXXXX
```
Once the configuration is complete, it is possible to send messages.
For now, we have found it helpful to embed the slacks in the `tryCatch` function.
```{r}
#| eval: false
tryCatch(
CODE YOU WANT TO RUN,
error = function(e)
{
#message to send if the code doesn't run
my_message <- paste( "example message")
slackr_msg(my_message, channel = "#recruitment")
})
```
## Slacking Specific Users
It is also possible to slack specific users with instructions found [at this link](https://stackoverflow.com/questions/32419756/how-do-you-tag-people-with-a-slack-bot) (archived at <https://perma.cc/59U5-V4GQ>).
# Replacing `//n` with a space
Many notes in projects that are exported from REDCap come with spaces denotes as `//n`.
Use the below code to make these fields more readable in the future.
```{r}
#| eval: false
gsub('\\n', ', ', df$notesField)
```
# Working with `R` on a Network Drive
When working with `R` on a network drive, it may be helpful to configure the project to store `.Rproj.user` on the local `C:/` drive rather than on the network drive, which results in slow execution times.
For more info:
- <https://github.com/rstudio/rstudio/pull/14875>
- <https://github.com/rstudio/rstudio/issues/14619#issuecomment-2189855598>
- <https://github.com/rstudio/rstudio/issues/10417#issuecomment-2199930385>
# Package Development
## Working with `renv` for Package Management
`renv` is used for reproducibility, by helping with package management (tracking package versions, etc.):
<https://rstudio.github.io/renv/articles/renv.html>
Before updating a package locally, make sure that it is available in the Posit Package Manager (so it can be available to GitHub Actions):
<https://packagemanager.posit.co>
### Updating the Package
1. To update the package, run the following in `R`:
```{r}
#| eval: false
# 1. Update packages in package environment
renv::upgrade()
renv::update()
renv::snapshot()
# 2. Add/edit code
# 3. Update documentation
roxygen2::roxygenise()
# 4. Update package version (or can do this manually)
usethis::use_version()
```
1. Then, build the package: Ctrl-Shift-B
1. Then, check the package: Ctrl-Shift-E
1. Then, run [`R CMD check`](#sec-rCmdCheck)
1. Then, install the package:
```{r}
#| eval: false
renv::install("C:/R/Packages/petersenlab") #PC
renv::install("/Library/Frameworks/R.framework/Packages/petersenlab") #Mac
```
### Installing Packages
To install new packages in the package environment, run the following in `R`:
```{r}
#| eval: false
renv::install("NAME_OF_PACKAGE")
```
or:
```{r}
#| eval: false
install.packages("NAME_OF_PACKAGE")
```
## `R CMD check` {#sec-rCmdCheck}
1. Build the source package (`.tar.gz` file)
- Click on the "Build" tab in the top-right pane of RStudio, and then click "Build Source Package", or run the following code in the `R` console:
```{r}
#| eval: false
devtools::build(pkg = "D:/Documents/GitHub/petersenlab")
```
1. Open terminal in RStudio
- After the package source (`.tar.gz` file) is built, open a terminal window directly in RStudio by clicking on the "Terminal" tab at the bottom of RStudio
1. Run `R CMD check --as-cran`
- In the terminal window, navigate to the directory where your package source (`.tar.gz` file) is located (commonly up one directory):
```bash
cd ..
```
Then, run `R CMD check --as-cran` followed by the name of your package tarball (making sure to update the package version in the filename). For example:
```bash
R CMD check --as-cran petersenlab_1.0.0.tar.gz
```
### Troubleshooting
#### If errors compiling the PDF manual
1. In the terminal:
```bash
cd ./petersenlab
R CMD Rd2pdf . --output=man/figures/manual.pdf --force --no-preview --no-clean
```
1. Then, delete the created folder whose name begins with ".Rd2pdf..."
1. In the R Console, build the source package in `R` (or using the instructions described [above](#sec-rCmdCheck)):
```{r}
#| eval: false
devtools::build(pkg = "D:/Documents/GitHub/petersenlab")
```
1. In the terminal (making sure to update the package version in the filename):
```bash
cd ..
R CMD check --as-cran petersenlab_1.0.0.tar.gz
```
#### `no visible binding for global variable`; `Undefined global functions or variables`
For example:
```r
no visible binding for global variable
'moderatorVal_centered'
Undefined global functions or variables:
moderatorVal_centered predictorVal_centered
```
Solution: set each variable to `NULL` in the package function before it is mentioned.
For example:
```r
predictorVal_centered <- moderatorVal_centered <- NULL
```
## `R CMD check` via GitHub Actions
```{r}
#| eval: false
usethis::use_github_action("check-standard")
```
## Useful keyboard shortcuts for package authoring:
Install Package: 'Ctrl + Shift + B'
Check Package: 'Ctrl + Shift + E'
Test Package: 'Ctrl + Shift + T'
```{r}
#| eval: false
renv::install("C:/R/Packages/petersenlab")
renv::snapshot()
renv::install()
```
## `pkgdown`
Run once to configure your package to use pkgdown:
```{r}
#| eval: false
usethis::use_pkgdown()
```
Then use `pkgdown` to build your website:
```{r}
#| eval: false
pkgdown::build_site()
```
## Steps to Add Functions
1. Add `.R` file with the function
1. Add the function to the `_pkgdown.yml` file
1. Update version number
1. `renv::upgrade()`
1. `renv::update()`
1. `renv::snapshot()`
1. `roxygen2::roxygenise()`
1. Install Package: 'Ctrl + Shift + B'
1. Check Package: 'Ctrl + Shift + E'
1. `R CMD check`
1. Commit and push changes
1. Update release version in GitHub
## Add sub-packages
```{r}
#| eval: false
devtools::build(pkg = "D:/Documents/GitHub/petersenlab/inst/extdata/testpackage1")
devtools::build(pkg = "D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2")
install.packages("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage1_0.1.0.tar.gz", repos = NULL, source = TRUE)
install.packages("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2_0.1.0.tar.gz", repos = NULL, source = TRUE)
remotes::install_local("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2_0.1.0.tar.gz")
remotes::install_local("D:/Documents/GitHub/petersenlab/inst/extdata/testpackage2_0.1.0.tar.gz")
```
## Submit Package to CRAN
<https://cran.r-project.org/submit.html>
## Resources
Official documentation for CRAN:
- <https://cran.r-project.org/doc/manuals/R-exts.html>
Unofficial documentation:
- <https://rpubs.com/YaRrr/rpackageintro>
- <https://r-pkgs.org>
- <https://cran.r-project.org/doc/contrib/Leisch-CreatingPackages.pdf>
- <https://portal.stats.ox.ac.uk/userdata/ruth/APTS2012/Rcourse10.pdf>
- <https://web.mit.edu/insong/www/pdf/rpackage_instructions.pdf>
- <https://hilaryparker.com/2014/04/29/writing-an-r-package-from-scratch/>
### For Package Development Tasks
- <https://usethis.r-lib.org/index.html>
### For Documentation
- <https://github.com/r-lib/roxygen2#roxygen2>
- <https://hilaryparker.com/2014/04/29/writing-an-r-package-from-scratch/>
### Creating Vignettes
- <https://bookdown.org/yihui/rmarkdown-cookbook/package-vignette.html>
### For Licensing
- <https://usethis.r-lib.org/reference/licenses.html>
# Troubleshooting {#sec-troubleshootingR}
To troubleshoot `R` issues, be resourceful.
Try googling the error message or issue you are experiencing.
See [here](#sec-questions) for a list of places you can pose `R`-related questions for help.
In addition, particular errors/warnings/issues are included below:
## General Troubleshooting Tips
1. **Run code line-by-line to identify the source of the error**
- If you are using RStudio, a red line will often appear on the lefthand side of the screen to indicate the particular line(s) of code that result in an error
- It may be useful to clear your working environment in R before trying to diagnose an error to be sure that you are not experiencing any interference from existing objects in the environment
- Running code line-by-line is particularly useful because it allows you to inspect the data at each step of its processing
- Data issues are commonly the culprit of code errors (see below)
1. **Check for typos**
- Are all brackets/parentheses/etc. closed?
- Does the object/variable that you are referring to exist? (*If the object does not exisit, you will likely get an error message indicating `[object name] not found`*)
- Has it been defined somewhere prior in the code?
- Does it depend on something else to be created?
- Is its name spelled correctly?
- Highlighting the object or variable in question and using `CTRL-F` to search for it is a good way to check these things
- Did you spell the function you want to use correctly?
- Do you have commas separating items within a list/function/etc.?
1. **Does your working environment have what it needs?**
- Are all necessary packages/libraries installed?
Do any require an update?
- Are all expected objects (dataframes, lists, environment variables, etc) present?
Are they correct?
1. **Investigate the structure of your data**
- Errors such as `non-numeric argument to binary operator` indicate that the function you are trying to use is expecting to recieve numeric data as an input but one or more of the inputs are not numeric
- The function `class(data$variable)` (replace "data" and "variable" with whatever you are trying to invesitgate) is useful for determining how R is interpreting your data.
Often, data that look like numbers can end up being stored as a character (text) variable.
This can be due to import/export processes or due to an issue with data entry.
- For example, when `00` is entered instead of `0` in a numeric field R is likely to interpret the variable as a character instead of a number when importing the data
- Check to make sure that all expected rows/columns are present and that they look the way you expect them to
- If your dataset has been created by joining/merging multiple dataframes, any duplicate columns not accounted for when joining may have a suffix appended to distinguish them (i.e., `variable.x` or `variable.y`)
- To resolve this, either call the appended variable in subsequent manipulations (`variable.x` instead of `variable`) or deal with the duplicated columns before/during the joining process (e.g., include duplicate columns in the `by` argument of joining function or remove the redundant column from one of the datasets to be joined)
- If you have been creating/computing variables in your dataset, ensure that they are being computed as expected
- Check for `NA` (missing) or `NaN` (not a number) values.
Such values may not be an issue or error in all cases, but if you are not expecting that as a result of your computations, this might suggest an issue with the code and/or data structure
- Check whether the computed values are reasonable
## A single error preventing a Wiki site from rendering
Code can be wrapped in `try()` statements so that if an error occurs, the script will simply "skip" to the next line of code.
These `try()` statements are particularly useful when rendering a site; if they are not used, a single error will halt the entire process. `try()` statements allow the code to continue running in spite of a line or two erroring out.
If you have already incorporated `try()` statements and are still experiencing fatal errors in code, check to make sure that the smallest possible "unit" of code is wrapped in `try()` statements, rather than large sections of code.
See [here](#sec-try) for more information.
## Warning: `PACKAGENAME` package in `FILEPATH` library will not be updated
This warning might indicate that it cannot update the package because it is part of the base `R` installation.
To fix this:
- find and delete the package that is part of the base `R` installation in the `library` directory of the `R` installation (e.g., `C:\R\R-4.3.1\library\PACKAGENAME\`)
- make sure the package is also not in any other package installation locations (e.g., `C:\R\Packages\PACKAGENAME\`); if it is, delete it from there as well
- then, after deleting the package from these locations, restart `R` and run `install.packages("PACKAGENAME")` to reinstall the package
- close `R`
- if the package was part of the base `R` installation, move the installed package folder `C:\R\Packages\PACKAGENAME\` to the `library` directory of the `R` installation (e.g., `C:\R\R-4.3.1\library\PACKAGENAME\`)
# Session Info
```{r}
#| code-fold: true
sessionInfo()
```
7.5 Comment code frequently and clearly!
It is important to comment code frequently and clearly. You want you (and others) to easily be able to understand your code if you come back to it several years later!