Generates a plot of a 2-way interaction.
Usage
plot2WayInteraction(
predictor,
outcome,
moderator,
predictorLabel = "predictor",
outcomeLabel = "outcome",
moderatorLabel = "moderator",
varList,
varTypes,
values = NA,
interaction = "normal",
legendLabels = NA,
legendLocation = "topright",
ylim = NA,
pvalues = TRUE,
data
)Arguments
- predictor
character name of predictor variable (variable on x-axis).
- outcome
character name of outcome variable (variable on y-axis).
- moderator
character name of moderator variable (variable on z-axis).
- predictorLabel
label on x-axis of plot
- outcomeLabel
label on y-axis of plot
- moderatorLabel
label on z-axis of plot
- varList
names of predictor variables in model
- varTypes
types of predictor variables in model; one of:
"mean"= plots at mean of variable – should be used for ALL covariates (apart from main predictor and moderator)"sd"= plots at +/- 1 sd of variable (for most continuous predictors and moderators)"binary"= plots at values of 0,1 (for binary predictors and moderators)"full"= plots full range of variable (for variables like age when on x-axis)"values"= allows plotting moderator at specific values (e.g.,0,1,2)"factor"= plots moderator at different categories (e.g.,TRUE,FALSE)
- values
specifies values at which to plot moderator (must specify varType =
"values")- interaction
one of:
"normal"= standard interaction"meancenter"= calculates the interaction from a mean-centered predictor and moderator (subtracting each individual's value from the variable mean to set the mean of the variable to zero)"orthogonalize"= makes the interaction orthogonal to the predictor and moderator by regressing the interaction on the predictor and outcome and saving the residual
- legendLabels
vector of 2 labels for the two levels of the moderator; leave as
NAto see the actual levels of the moderator- legendLocation
one of:
"topleft","topright","bottomleft", or"bottomright"- ylim
vector of min and max values on y-axis (e,g.,
0,10)- pvalues
whether to include p-values of each slope in plot (
TRUEorFALSE)- data
name of data object
Details
Generates a plot of a 2-way interaction: the association between a predictor and an outcome at two levels of the moderator.
See also
Other plot:
addText(),
ppPlot(),
semPlotInteraction(),
vwReg()
Other multipleRegression:
lmCombine(),
ppPlot(),
semPlotInteraction(),
update_nested()
Examples
# Prepare Data
predictor <- rnorm(1000, 10, 3)
moderator <- rnorm(1000, 50, 10)
outcome <- (1.7 * predictor) + (1.3 * moderator) +
(1.5 * predictor * moderator) + rnorm(1000, sd = 3)
covariate <- rnorm(1000)
df <- data.frame(predictor, moderator, outcome, covariate)
# Linear Regression
lmModel <- lm(outcome ~ predictor + moderator + predictor:moderator,
data = df, na.action = "na.exclude")
summary(lmModel)
#>
#> Call:
#> lm(formula = outcome ~ predictor + moderator + predictor:moderator,
#> data = df, na.action = "na.exclude")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3264 -2.0181 0.0738 1.9260 9.9847
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.731043 1.911170 1.952 0.0512 .
#> predictor 1.342935 0.184102 7.294 6.11e-13 ***
#> moderator 1.227713 0.037473 32.763 < 2e-16 ***
#> predictor:moderator 1.507044 0.003615 416.903 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.008 on 996 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.814e+06 on 3 and 996 DF, p-value: < 2.2e-16
#>
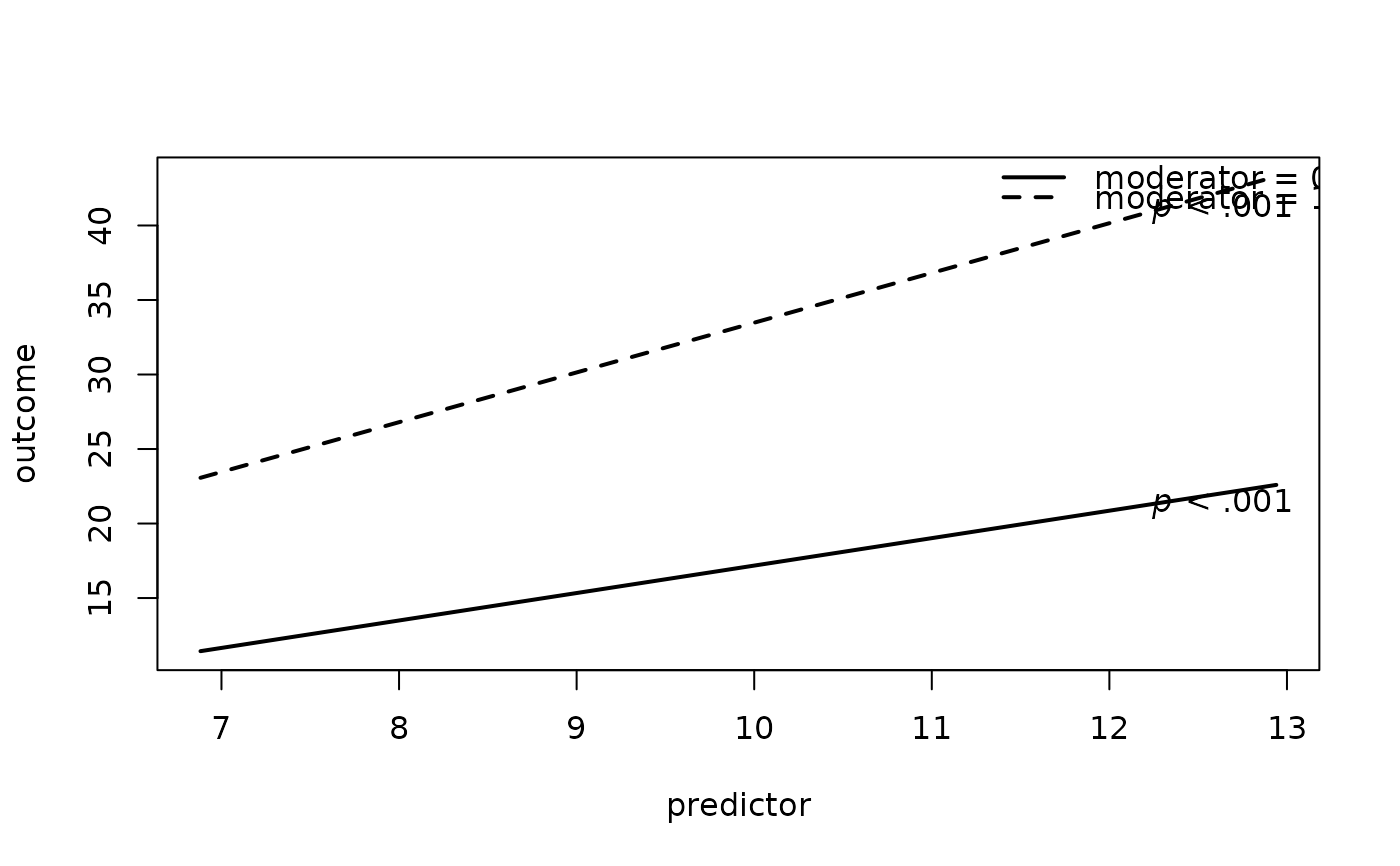
# 1. Plot 2-Way Interaction
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
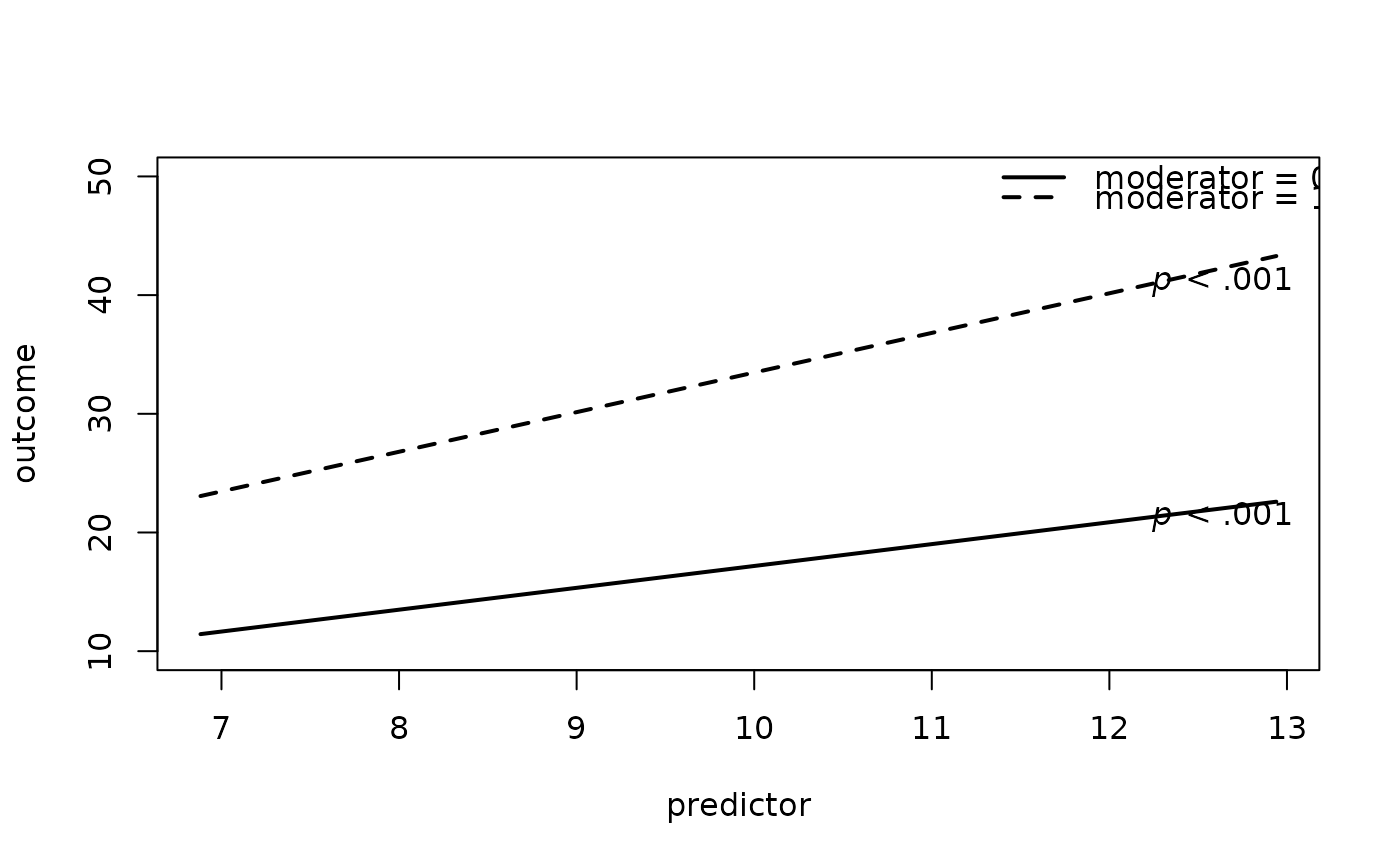
# 2. Specify y-axis Range
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50), #new
data = df)
#>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
# 2. Specify y-axis Range
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50), #new
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
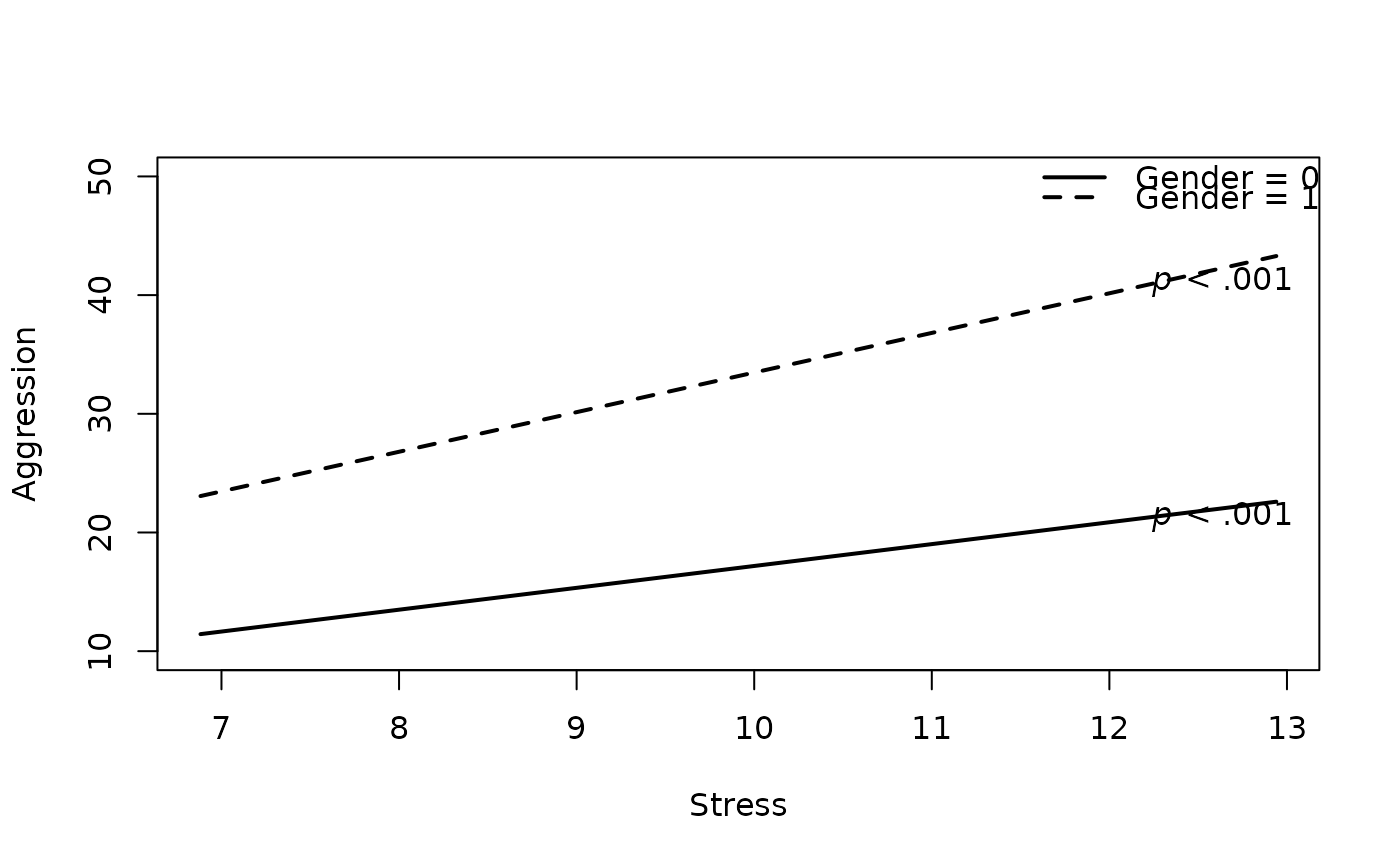
# 3. Add Variable Labels
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress", #new
outcomeLabel = "Aggression", #new
moderatorLabel = "Gender", #new
data = df)
#>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
# 3. Add Variable Labels
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress", #new
outcomeLabel = "Aggression", #new
moderatorLabel = "Gender", #new
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
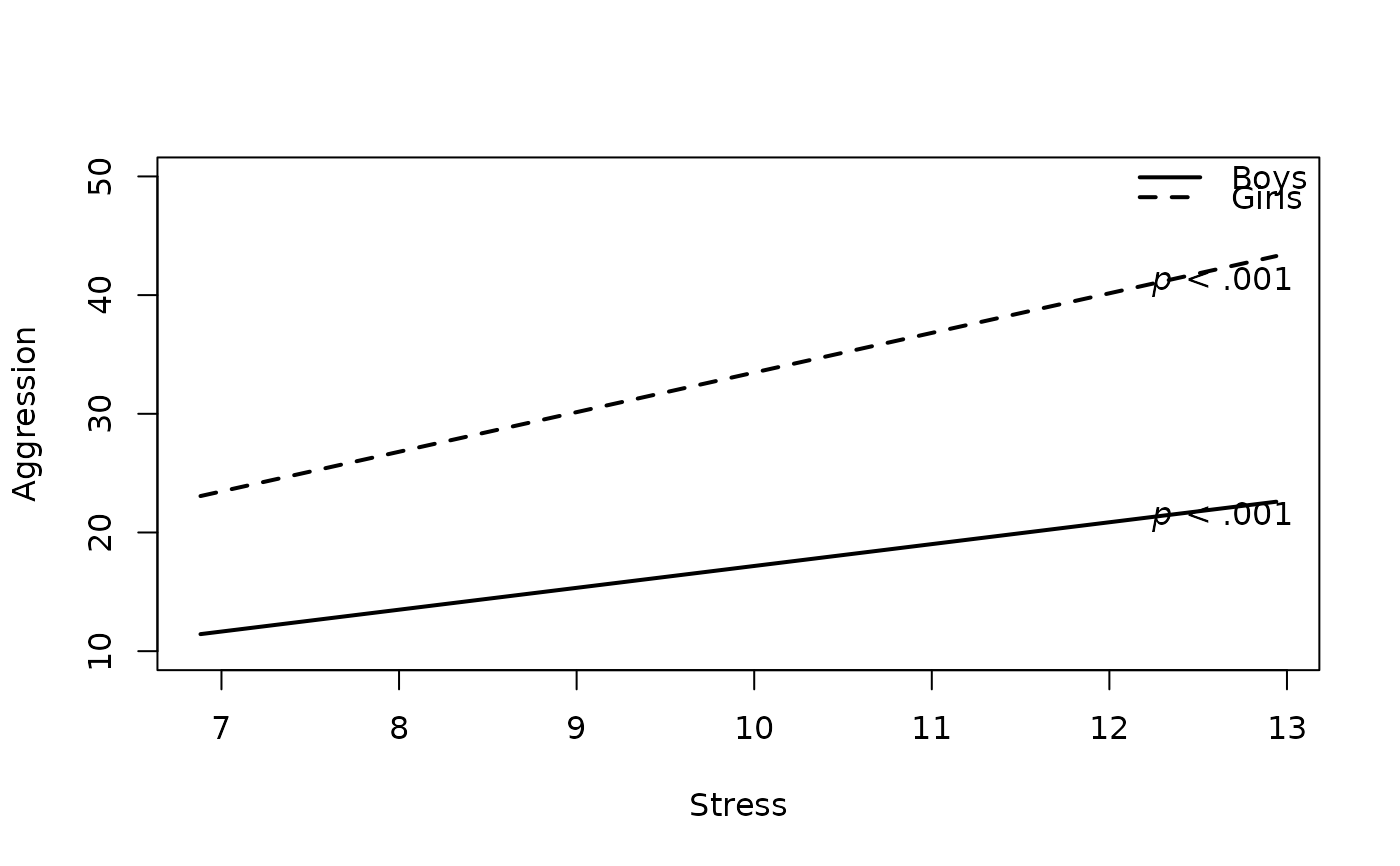
# 4. Change Legend Labels
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"), #new
data = df)
#>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
# 4. Change Legend Labels
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"), #new
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
# 5. Move Legend Location
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft", #new
data = df)
#>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
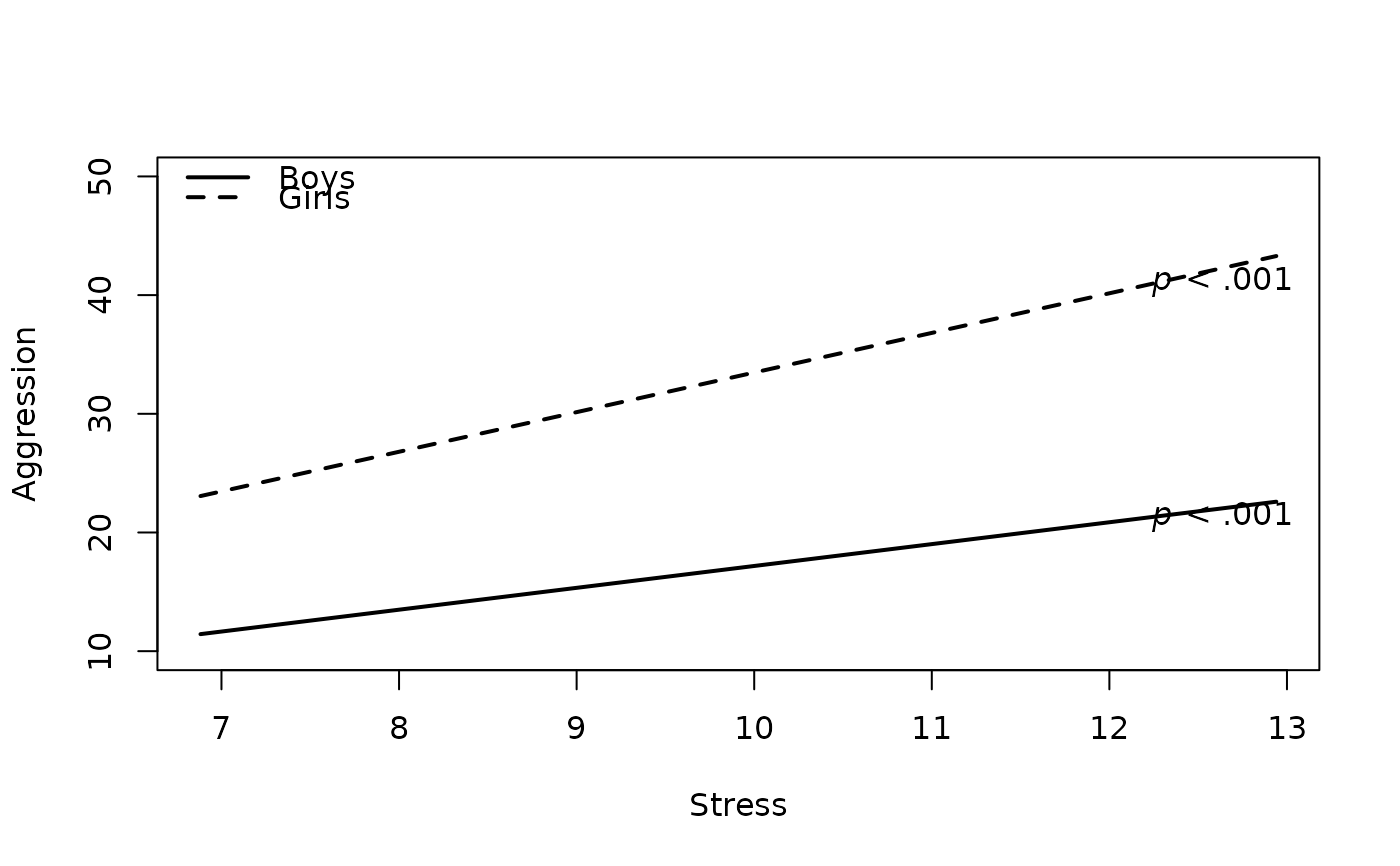
# 5. Move Legend Location
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft", #new
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
#6. Turn Off p-Values
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft",
pvalues = FALSE, #new
data = df)
#>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
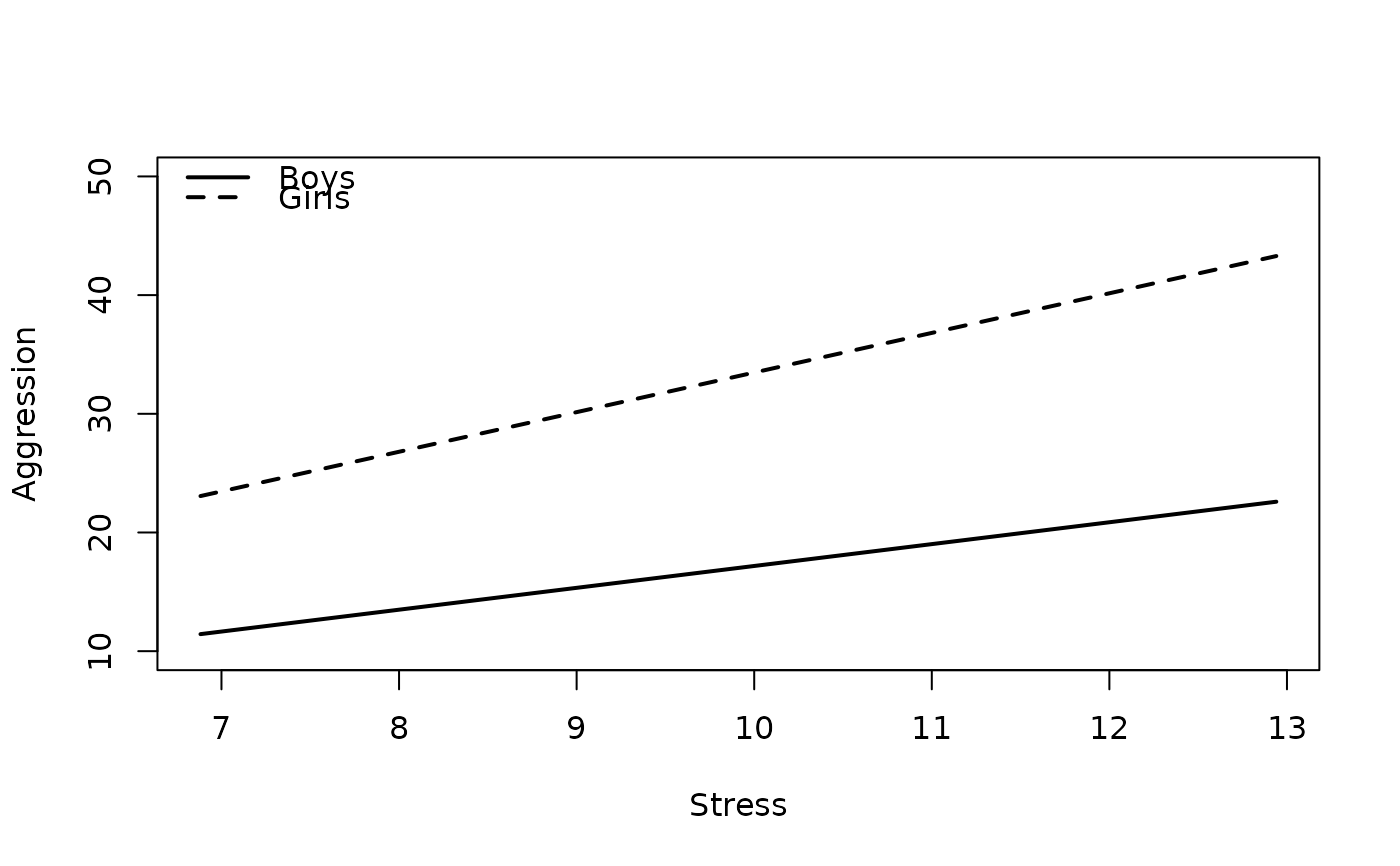
#6. Turn Off p-Values
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft",
pvalues = FALSE, #new
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
#7. Get Regression Output from Mean-Centered Predictor and Moderator
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft",
interaction = "meancenter", #new
data = df)
#>
#> Call:
#> lm(formula = modelFormula, data = modelData, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.728853 1.912453 1.950 0.0515 .
#> predictor 1.343106 0.184215 7.291 6.27e-13 ***
#> moderator 1.227771 0.037503 32.738 < 2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.9506
#> predictor:moderator 1.507039 0.003618 416.595 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
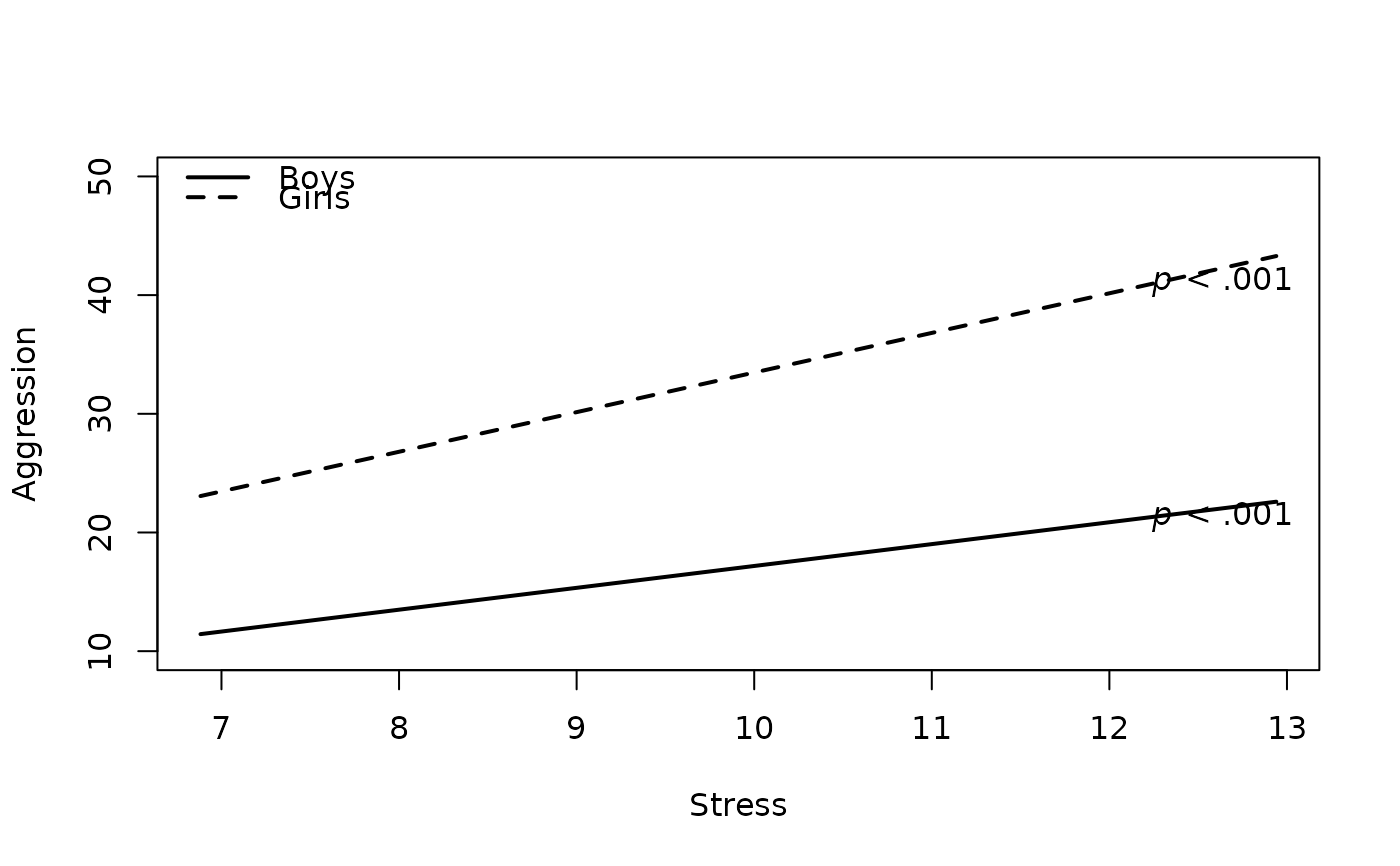
#7. Get Regression Output from Mean-Centered Predictor and Moderator
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft",
interaction = "meancenter", #new
data = df)
 #>
#> Call:
#> lm(formula = modelFormula, data = modelDataMC, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 837.833289 0.095178 8802.779 <2e-16 ***
#> predictor 76.931990 0.032615 2358.771 <2e-16 ***
#> moderator 16.360910 0.009773 1674.108 <2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.951
#> predictor:moderator 1.507039 0.003618 416.595 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
#8. Get Regression Output from Orthogonalized Interaction Term
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft",
interaction = "orthogonalize", #new
data = df)
#>
#> Call:
#> lm(formula = modelFormulaOrtho, data = modelDataOrtho, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -7.528e+02 6.001e-01 -1254.453 <2e-16 ***
#> predictor 7.688e+01 3.261e-02 2357.335 <2e-16 ***
#> moderator 1.631e+01 9.772e-03 1669.050 <2e-16 ***
#> covariate -5.692e-03 9.181e-02 -0.062 0.951
#> predictorXmoderator 1.507e+00 3.618e-03 416.595 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
#>
#> Call:
#> lm(formula = modelFormula, data = modelDataMC, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 837.833289 0.095178 8802.779 <2e-16 ***
#> predictor 76.931990 0.032615 2358.771 <2e-16 ***
#> moderator 16.360910 0.009773 1674.108 <2e-16 ***
#> covariate -0.005692 0.091811 -0.062 0.951
#> predictor:moderator 1.507039 0.003618 416.595 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
#8. Get Regression Output from Orthogonalized Interaction Term
plot2WayInteraction(predictor = "predictor",
outcome = "outcome",
moderator = "moderator",
varList = c("predictor","moderator","covariate"),
varTypes = c("sd","binary","mean"),
ylim = c(10,50),
predictorLabel = "Stress",
outcomeLabel = "Aggression",
moderatorLabel = "Gender",
legendLabels = c("Boys","Girls"),
legendLocation = "topleft",
interaction = "orthogonalize", #new
data = df)
#>
#> Call:
#> lm(formula = modelFormulaOrtho, data = modelDataOrtho, na.action = "na.omit")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -10.3153 -2.0199 0.0733 1.9282 9.9801
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -7.528e+02 6.001e-01 -1254.453 <2e-16 ***
#> predictor 7.688e+01 3.261e-02 2357.335 <2e-16 ***
#> moderator 1.631e+01 9.772e-03 1669.050 <2e-16 ***
#> covariate -5.692e-03 9.181e-02 -0.062 0.951
#> predictorXmoderator 1.507e+00 3.618e-03 416.595 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 3.009 on 995 degrees of freedom
#> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
#> F-statistic: 2.108e+06 on 4 and 995 DF, p-value: < 2.2e-16
#>
